ClusterDepth
Fast implementation (~>100x faster than R/C++) of the ClusterDepth multiple comparison algorithm from Frossard and Renaud Neuroimage 2022
This is especially interesting to EEG signals. Currently only acts on a single channel/timeseries. Multichannel as discussed in the paper is the next step.
Quickstart
using ClusterDepth
pval_corrected = clusterdepth(erpMatrix; τ=2.3,nperm=5000)
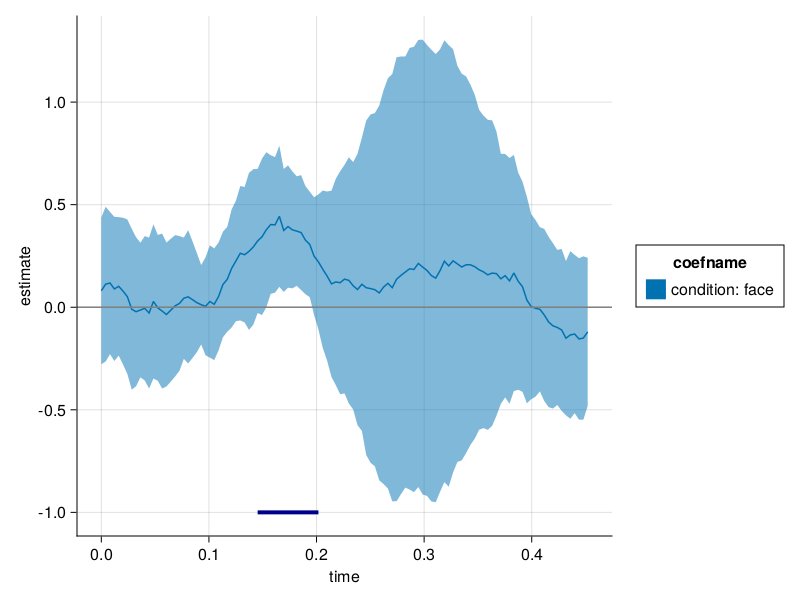
FWER check
We checked FWER for troendle(...) and clusterdepth(...) (link to docs)
For clustedepth we used 5000 repetitions, 5000 permutations, 200 tests.
| simulation | noise | uncorrected | type |
|---|---|---|---|
| clusterdepth | white | 1.0 | 0.0554 |
| clusterdepth | red* | 0.835 | 0.0394 |
| troendle | white | XX | XX |
| troendle | red* | XX | XX |
Uncorrected should be 1 - it is very improbable, than none of the 200 tests in one repetition is not significant (we expect 5% to be).
Corrected should be 0.05 (CI-95 [0.043,0.0564])
* red noise introduces strong correlations between individual trials, thus makes the tests correlated while following the H0.
Citing
Algorithm published in https://doi.org/10.1016/j.neuroimage.2021.118824 - Frossard & Renaud 2022, Neuroimage
Please also cite this toolbox:
Some functions are inspired by R::permuco, written by Jaromil Frossard. Note: Permuco is GPL licensed, but Jaromil Frossard released the relevant clusteterdepth functions to me under MIT. Thereby, this repository can be licensed under MIT.
1
