TopicModelsVB.jl
v1.x compatible.
A Julia package for variational Bayesian topic modeling.
Topic models are Bayesian hierarchical models designed to discover the latent low-dimensional thematic structure within corpora. Like most probabilistic graphical models, topic models are fit using either Markov chain Monte Carlo (MCMC), or variational inference (VI).
Markov chain Monte Carlo is slow but consistent, given infinite time MCMC will fit the desired model exactly. Unfortunately, the lack of an objective metric for assessing convergence means that it's difficult to state unequivocally that MCMC has reached an optimal steady-state.
Contrarily, variational inference is fast but inconsistent, since one must approximate distributions in order to ensure tractability. Fortunately, VI may be characterized in the language of numerical optimization, and its performance evaluated objectively via the assessment of convergence to local optima.
This package takes the latter approach to topic modeling.
Dependencies
DelimitedFiles
SpecialFunctions
LinearAlgebra
Random
Distributions
OpenCL
Crayons
Install
(@v1.5) pkg> add TopicModelsVB
Datasets
Included in TopicModelsVB.jl are two datasets:
- National Science Foundation Abstracts 1989 - 2003:
- 128804 documents
- 25319 vocabulary
- CiteULike Science Article Database:
- 16980 documents
- 8000 vocabulary
- 5551 users
Corpus
Let's begin with the Corpus data structure. The Corpus data structure has been designed for maximum ease-of-use. Datasets must still be cleaned and put into the appropriate format, but once a dataset is in the proper format and read into a corpus, it can easily be modified to meet the user's needs.
There are four plaintext files that make up a corpus:
- docfile
- vocabfile
- userfile
- titlefile
None of these files are mandatory to read a corpus, and in fact reading no files will result in an empty corpus. However in order to train a model a docfile will be necessary, since it contains all quantitative data known about the documents. On the other hand, the vocab, user and title files are used solely for interpreting output.
The docfile should be a plaintext file containing lines of delimited numerical values. Each document is a block of lines, the number of which depends on what information is known about the documents. Since a document is at its essence a list of terms, each document must contain at least one line containing a nonempty list of delimited positive integer values corresponding to the terms of which it is composed. Any further lines in a document block are optional, however if they are present they must be present for all documents and must come in the following order:
terms - A line of delimited positive integers corresponding to the terms which make up the document (this line is mandatory).
counts - A line of delimited positive integers, equal in length to terms, corresponding to the number of times a term appears in a document.
readers - A line of delimited positive integers corresponding to those users which have read the document.
ratings - A line of delimited positive integers, equal in length to readers, corresponding to the rating each reader gave the document.
An example of a single doc block from a docfile with all possible lines included,
...
4,10,3,100,57
1,1,2,1,3
1,9,10
1,1,5
...
The vocab and user files are tab delimited dictionaries mapping positive integers to terms and usernames (resp.). For example,
1 this
2 is
3 a
4 vocab
5 file
A userfile is identitcal to a vocabfile, except usernames will appear in place of vocabulary terms.
Finally, a titlefile is simply a list of titles, not a dictionary, and is of the form,
title1
title2
title3
title4
title5
The order of these titles correspond to the order of document blocks in the associated docfile.
To read a corpus into Julia, use the following function,
readcorp(;docfile="", vocabfile="", userfile="", titlefile="", delim=',', counts=false, readers=false, ratings=false)
The file keyword arguments indicate the path where the respective file is located.
It is often the case that even once files are correctly formatted and read, the corpus will still contain formatting defects which prevent it from being loaded into a model. Therefore, before loading a corpus into a model, it is important that one of the following is run,
fixcorp!(corp)
or
fixcorp!(corp, pad=true)
Padding a corpus will ensure that any documents which contain vocab or user keys not in the vocab or user dictionaries are not removed. Instead, generic vocab and user keys will be added as necessary to the vocab and user dictionaries (resp.).
The fixcorp! function allows for significant customization of the corpus object.
For example, let's begin by loading the CiteULike corpus,
corp = readcorp(:citeu)
A standard preprocessing step might involve removing stop words, removing terms which appear less than 200 times, and alphabetizing our corpus.
fixcorp!(corp, stop=true, abridge=200, alphabetize=true, trim=true)
### Generally you will also want to trim your corpus.
### Setting trim=true will remove leftover terms from the corpus vocabulary.
After removing stop words and abridging our corpus, the vocabulary size has gone from 8000 to 1692.
A consequence of removing so many terms from our corpus is that some documents may now by empty. We can remove these documents from our corpus with the following command,
fixcorp!(corp, remove_empty_docs=true)
In addition, if you would like to preserve term order in your documents, then you should refrain from condesing your corpus.
For example,
corp = Corpus(Document(1:9), vocab=split("the quick brown fox jumped over the lazy dog"))
showdocs(corp)
●●● Document 1
the quick brown fox jumped over the lazy dog
fixcorp!(corp, condense=true)
showdocs(corp)
●●● Document 1
the fox dog quick brown jumped lazy over the
Important. A corpus is only a container for documents.
Whenever you load a corpus into a model, a copy of that corpus is made, such that if you modify the original corpus at corpus-level (remove documents, re-order vocab keys, etc.), this will not affect any corpus attached to a model. However! Since corpora are containers for their documents, modifying an individual document will affect it in all corpora which contain it. Therefore,
Using
fixcorp!to modify the documents of a corpus will not result in corpus defects, but will cause them also to be changed in all other corpora which contain them.If you would like to make a copy of a corpus with independent documents, use
deepcopy(corp).Manually modifying documents is dangerous, and can result in corpus defects which cannot be fixed by
fixcorp!. It is advised that you don't do this without good reason.
Models
The available models are as follows:
CPU Models
LDA(corp, K)
Latent Dirichlet allocation model with K topics.
fLDA(corp, K)
Filtered latent Dirichlet allocation model with K topics.
CTM(corp, K)
Correlated topic model with K topics.
fCTM(corp, K)
Filtered correlated topic model with K topics.
CTPF(corp, K)
Collaborative topic Poisson factorization model with K topics.
GPU Models
gpuLDA(corp, K)
GPU accelerated latent Dirichlet allocation model with K topics.
gpuCTM(corp, K)
GPU accelerated correlated topic model with K topics.
gpuCTPF(corp, K)
GPU accelerated collaborative topic Poisson factorization model with K topics.
Tutorial
Latent Dirichlet Allocation
Let's begin our tutorial with a simple latent Dirichlet allocation (LDA) model with 9 topics, trained on the first 5000 documents from the NSF corpus.
using TopicModelsVB
using Random
using Distributions
Random.seed!(10);
corp = readcorp(:nsf)
corp.docs = corp[1:5000];
fixcorp!(corp, trim=true)
### It's strongly recommended that you trim your corpus when reducing its size in order to remove excess vocabulary.
### Notice that the post-fix vocabulary is smaller after removing all but the first 5000 docs.
model = LDA(corp, 9)
train!(model, iter=150, tol=0)
### Setting tol=0 will ensure that all 150 iterations are completed.
### If you don't want to compute the ∆elbo, set checkelbo=Inf.
### training...
showtopics(model, cols=9, 20)
topic 1 topic 2 topic 3 topic 4 topic 5 topic 6 topic 7 topic 8 topic 9
plant research models research data research research research theory
cell chemistry research project research system dr students problems
protein study study study species systems university science study
cells high data data study design support program research
genetic chemical model social project data award university equations
gene studies numerical theory important project program conference work
molecular surface theoretical economic provide earthquake sciences support geometry
studies materials methods understanding studies performance project scientists project
proteins metal problems important time control months provide groups
dna reactions theory work field based mathematical engineering algebraic
plants properties physics information ocean computer professor workshop differential
genes organic work development water analysis year faculty investigator
research program systems policy analysis algorithms science graduate space
study electron flow models understanding parallel equipment national principal
specific phase analysis behavior determine developed scientists scientific mathematical
system structure time provide results techniques institute international systems
important temperature processes analysis climate information scientific undergraduate analysis
function molecular solar political patterns time collaboration held spaces
understanding systems large model large network projects projects problem
development project project public processes structures national project solutions
If you are interested in the raw topic distributions. For LDA and CTM models, you may access them via the matrix,
model.beta
### K x V matrix
### K = number of topics.
### V = number of vocabulary terms, ordered identically to the keys in model.corp.vocab.
Now that we've trained our LDA model we can, if we want, take a look at the topic proportions for individual documents.
For instance, document 1 has topic breakdown,
println(round.(topicdist(model, 1), digits=3))
### = [0.161, 0.0, 0.0, 0.063, 0.774, 0.0, 0.0, 0.0, 0.0]
This vector of topic weights suggests that document 1 is mostly about biology, and in fact looking at the document text confirms this observation,
showdocs(model, 1)
### Could also have done showdocs(corp, 1).
●●● Document 1
●●● CRB: Genetic Diversity of Endangered Populations of Mysticete Whales: Mitochondrial DNA and Historical Demography
commercial exploitation past hundred years great extinction variation sizes
populations prior minimal population size current permit analyses effects
differing levels species distributions life history...
Just for fun, let's consider one more document (document 25),
println(round.(topicdist(model, 25), digits=3))
### = [0.0, 0.0, 0.583, 0.0, 0.0, 0.0, 0.0, 0.0, 0.415]
showdocs(model, 25)
●●● Document 25
●●● Mathematical Sciences: Nonlinear Partial Differential Equations from Hydrodynamics
work project continues mathematical research nonlinear elliptic problems arising perfect
fluid hydrodynamics emphasis analytical study propagation waves stratified media techniques
analysis partial differential equations form basis studies primary goals understand nature
internal presence vortex rings arise density stratification due salinity temperature...
We see that in this case document 25 appears to be about mathematical physics, which corresponds precisely to topics 3 and 9.
Furthermore, if we want to, we can also generate artificial corpora by using the gencorp function.
Generating artificial corpora will in turn run the underlying probabilistic graphical model as a generative process in order to produce entirely new collections of documents, let's try it out,
Random.seed!(10);
artificial_corp = gencorp(model, 5000, laplace_smooth=1e-5)
### The laplace_smooth argument governs the amount of Laplace smoothing (defaults to 0).
artificial_model = LDA(artificial_corp, 9)
train!(artificial_model, iter=150, tol=0, checkelbo=10)
### training...
showtopics(artificial_model, cols=9)
topic 1 topic 2 topic 3 topic 4 topic 5 topic 6 topic 7 topic 8 topic 9
research models research protein research research data research theory
project study study plant system dr research students problems
data research chemistry cell systems university species program study
system data surface cells design support project science equations
design methods high dna data award study conference research
systems theoretical materials genetic earthquake program provide university work
study model chemical gene project project time support project
information numerical metal proteins program sciences important scientists groups
earthquake problems electron molecular developed months studies engineering geometry
theory theory studies plants control mathematical analysis provide differential
models physics properties studies based professor processes workshop algebraic
analysis analysis organic research techniques equipment climate faculty investigator
control work program genes performance science results graduate mathematical
work systems reactions important time scientists field national systems
performance flow phase system high year water scientific principal
Correlated Topic Model
For our next model, let's upgrade to a (filtered) correlated topic model (fCTM).
Filtering the correlated topic model will dynamically identify and suppress stop words which would otherwise clutter up the topic distribution output.
Random.seed!(10);
model = fCTM(corp, 9)
train!(model, tol=0, checkelbo=Inf)
### training...
showtopics(model, 20, cols=9)
topic 1 topic 2 topic 3 topic 4 topic 5 topic 6 topic 7 topic 8 topic 9
design materials economic species earthquake students chemistry theory cell
system flow social ocean data university reactions problems protein
systems temperature theory populations seismic science university equations cells
algorithms surface policy water soil support metal geometry gene
parallel phase political data damage program organic investigator plant
performance high public climate university scientists molecular algebraic proteins
based optical decision marine stars sciences chemical groups genes
networks laser labor sea buildings conference compounds principal dna
network properties market plant ground scientific molecules mathematical molecular
control liquid data population response national professor differential plants
computer measurements children patterns solar year reaction space genetic
processing experimental science evolutionary equipment engineering synthesis problem regulation
problems heat change plants nsf faculty program solutions expression
software growth people genetic national workshop electron mathematics growth
programming electron women north california mathematical complexes spaces specific
distributed films human pacific san months department nonlinear function
neural gas factors change program graduate energy finite binding
applications fluid groups samples hazard projects species manifolds cellular
efficient quantum individuals environmental earthquakes academic spectroscopy functions membrane
problem solid case history october international carbon dimensional sequence
Based on the top 20 terms in each topic, we might tentatively assign the following topic labels:
- topic 1: Computer Science
- topic 2: Physics
- topic 3: Economics
- topic 4: Ecology
- topic 5: Earthquakes
- topic 6: Academia
- topic 7: Chemistry
- topic 8: Mathematics
- topic 9: Molecular Biology
Now let's take a look at the topic-covariance matrix,
model.sigma
### Top two off-diagonal positive entries:
model.sigma[4,9] # = 11.219
model.sigma[1,8] # = 4.639
### Top two negative entries:
model.sigma[4,8] # = -34.815
model.sigma[8,9] # = -13.546
According to the list above, the most closely related topics are topics 4 and 9, which correspond to the Ecology and Molecular Biology topics, followed by 1 and 8, corresponding to Computer Science and Mathematics.
As for the most unlikely topic pairings, most strongly negatively correlated are topics 4 and 8, corresponding to Ecology and Mathematics, followed by topics 8 and 9, corresponding to Mathematics and Molecular Biology.
Topic Prediction
The topic models so far discussed can also be used to train a classification algorithm designed to predict the topic distribution of new, unseen documents.
Let's take our 5,000 document NSF corpus, and partition it into training and test corpora,
train_corp = copy(corp)
train_corp.docs = train_corp[1:4995];
test_corp = copy(corp)
test_corp.docs = test_corp[4996:5000];
Now we can train our LDA model on just the training corpus, and then use that trained model to predict the topic distributions of the five documents in our test corpus,
Random.seed!(10);
train_model = LDA(train_corp, 9)
train!(train_model, checkelbo=Inf)
test_model = predict(test_corp, train_model)
The predict function works by taking in a corpus of new, unseen documents, and a trained model, and returning a new model of the same type. This new model can then be inspected directly, or using topicdist, in order to see the topic distribution for the documents in the test corpus.
Let's first take a look at both the topics for the trained model and the documents in our test corpus,
showtopics(train_model, cols=9, 20)
topic 1 topic 2 topic 3 topic 4 topic 5 topic 6 topic 7 topic 8 topic 9
plant research models research data research research research theory
cell chemistry research project research system dr students problems
protein study study study species systems university science study
cells high data data study design support program research
genetic chemical model theory project data award university equations
gene studies numerical social important project program conference work
molecular surface methods economic provide earthquake sciences support geometry
studies materials theoretical understanding studies performance project scientists groups
proteins metal problems important time control months provide algebraic
dna reactions theory work field based mathematical engineering project
plants properties work information ocean computer professor workshop differential
genes organic physics development analysis analysis year faculty investigator
research program systems policy water algorithms science graduate space
study electron analysis models understanding parallel equipment national mathematical
specific phase flow provide determine information scientists scientific principal
system structure time behavior results techniques institute international systems
important temperature large analysis climate developed scientific undergraduate spaces
function molecular processes political patterns time collaboration held analysis
understanding systems solar model large network projects project solutions
development measurements project public processes structures national projects mathematics
showtitles(corp, 4996:5000)
• Document 4996 Decision-Making, Modeling and Forecasting Hydrometeorologic Extremes Under Climate Change
• Document 4997 Mathematical Sciences: Representation Theory Conference, September 13-15, 1991, Eugene, Oregon
• Document 4998 Irregularity Modeling & Plasma Line Studies at High Latitudes
• Document 4999 Uses and Simulation of Randomness: Applications to Cryptography,Program Checking and Counting Problems.
• Document 5000 New Possibilities for Understanding the Role of Neuromelanin
Now let's take a look at the predicted topic distributions for these five documents,
for d in 1:5
println("Document ", 4995 + d, ": ", round.(topicdist(test_model, d), digits=3))
end
Document 4996: [0.0, 0.001, 0.207, 0.188, 0.452, 0.151, 0.0, 0.0, 0.0]
Document 4997: [0.001, 0.026, 0.001, 0.043, 0.001, 0.001, 0.012, 0.386, 0.53]
Document 4998: [0.0, 0.019, 0.583, 0.0, 0.268, 0.122, 0.0, 0.007, 0.0]
Document 4999: [0.002, 0.002, 0.247, 0.037, 0.019, 0.227, 0.002, 0.026, 0.438]
Document 5000: [0.785, 0.178, 0.001, 0.0, 0.034, 0.001, 0.001, 0.001, 0.0]
Collaborative Topic Poisson Factorization
For our final model, we take a look at the collaborative topic Poisson factorization (CTPF) model.
CTPF is a collaborative filtering topic model which uses the latent thematic structure of documents to improve the quality of document recommendations beyond what would be possible using just the document-user matrix alone. This blending of thematic structure with known user prefrences not only improves recommendation accuracy, but also mitigates the cold-start problem of recommending to users never-before-seen documents. As an example, let's load the CiteULike dataset into a corpus and then randomly remove a single reader from each of the documents.
Random.seed!(10);
corp = readcorp(:citeu)
ukeys_test = Int[];
for doc in corp
index = sample(1:length(doc.readers), 1)[1]
push!(ukeys_test, doc.readers[index])
deleteat!(doc.readers, index)
deleteat!(doc.ratings, index)
end
Important. We refrain from fixing our corpus in this case, first because the CiteULike dataset is pre-packaged and thus pre-fixed, but more importantly, because removing user keys from documents and then fixing a corpus may result in a re-ordering of its user dictionary, which would in turn invalidate our test set.
After training, we will evaluate model quality by measuring our model's success at imputing the correct user back into each of the document libraries.
It's also worth noting that after removing a single reader from each document, 158 of the documents now have 0 readers,
sum([isempty(doc.readers) for doc in corp]) # = 158
Fortunately, since CTPF can if need be depend entirely on thematic structure when making recommendations, this poses no problem for the model.
Now that we've set up our experiment, let's instantiate and train a CTPF model on our corpus. Furthermore, in the interest of time, we'll also go ahead and GPU accelerate it.
model = gpuCTPF(corp, 100)
train!(model, iter=50, checkelbo=Inf)
### training...
Finally, we evaluate the performance of our model on the test set.
ranks = Float64[];
for (d, u) in enumerate(ukeys_test)
urank = findall(model.drecs[d] .== u)[1]
nrlen = length(model.drecs[d])
push!(ranks, (nrlen - urank) / (nrlen - 1))
end
The following histogram shows the proportional ranking of each test user within the list of recommendations for their corresponding document.
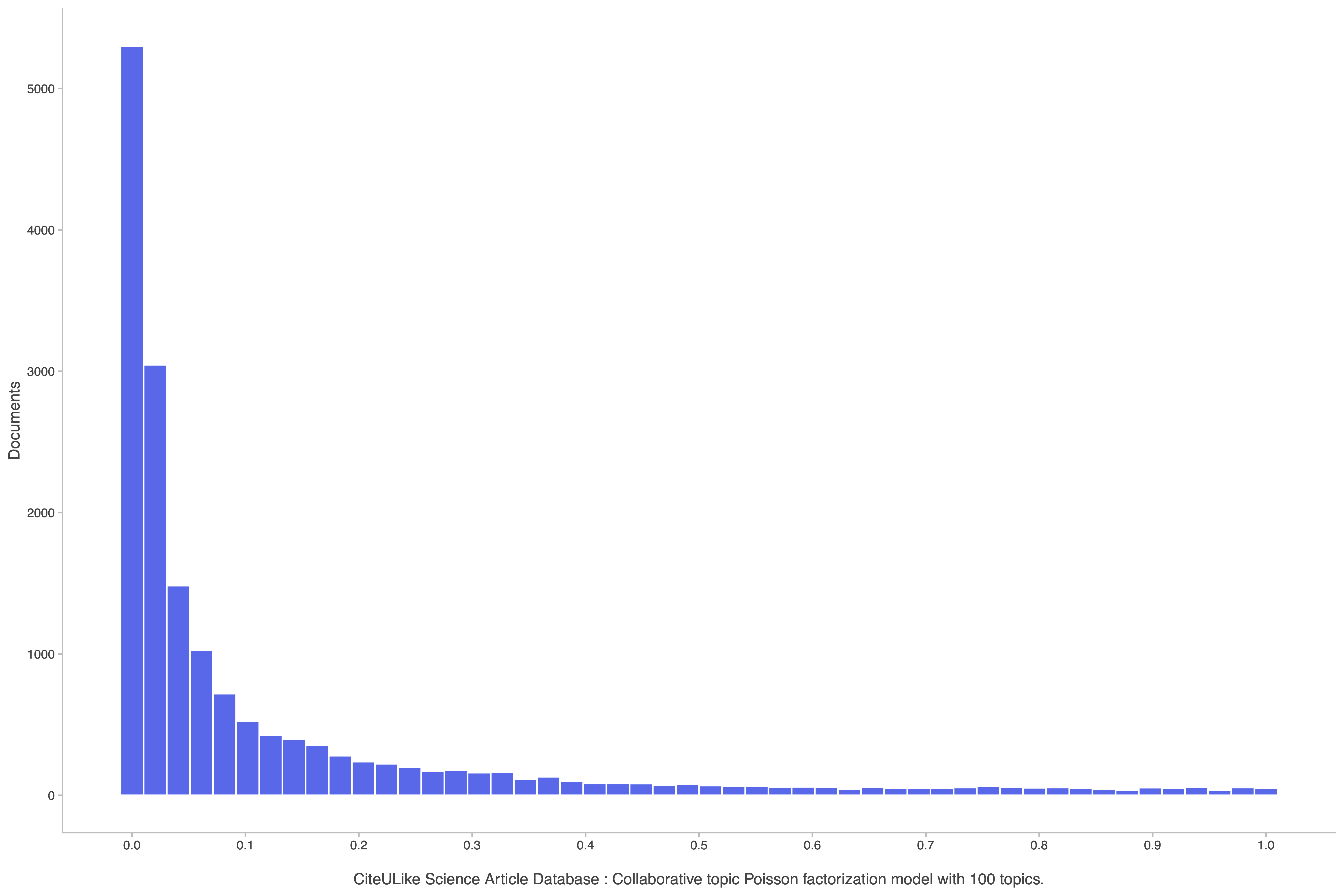
Let's also take a look at the top recommendations for a particular document,
ukeys_test[1] # = 4919
ranks[1] # = 0.958
showdrecs(model, 1, 231)
●●● doc 1
●●● The metabolic world of Escherichia coli is not small
...
228. #user4415
229. #user5167
230. #user429
231. #user4919
What the above output tells us is that user 4919's test document placed him or her in the top 4.2% (position 231) of all non-readers.
For evaluating our model's user recommendations, let's take a more holistic approach.
Since large heterogenous libraries make the qualitative assessment of recommendations difficult, let's search for a user with a small focused library,
showlibs(model, 1741)
●●● user 1741
• Region-Based Memory Management
• A Syntactic Approach to Type Soundness
• Imperative Functional Programming
• The essence of functional programming
• Representing monads
• The marriage of effects and monads
• A Taste of Linear Logic
• Monad transformers and modular interpreters
• Comprehending Monads
• Monads for functional programming
• Building interpreters by composing monads
• Typed memory management via static capabilities
• Computational Lambda-Calculus and Monads
• Why functional programming matters
• Tackling the Awkward Squad: monadic input/output, concurrency, exceptions, and foreign-language calls in Haskell
• Notions of Computation and Monads
• Recursion schemes from comonads
• There and back again: arrows for invertible programming
• Composing monads using coproducts
• An Introduction to Category Theory, Category Theory Monads, and Their Relationship to Functional Programming
The 20 articles in user 1741's library suggest that he or she is interested in programming language theory.
Now compare this with the top 50 recommendations (the top 0.3%) made by our model,
showurecs(model, 1741, 50)
●●● User 1741
1. Sets for Mathematics
2. On Understanding Types, Data Abstraction, and Polymorphism
3. Can programming be liberated from the von {N}eumann style? {A} functional style and its algebra of programs
4. Types and programming languages
5. Haskell's overlooked object system
6. Obol: integrating language and meaning in bio-ontologies
7. The Definition of Standard ML - Revised
8. Advanced Topics in Types and Programming Languages
9. Principles of programming with complex objects and collection types
10. Why Dependent Types Matter
11. Functional programming with bananas, lenses, envelopes and barbed wire
12. Featherweight Java: A Minimal Core Calculus for Java and GJ
13. On the expressive power of programming languages
14. Modern {C}ompiler {I}mplementation in {J}ava
15. Functional pearl: implicit configurations--or, type classes reflect the values of types
16. Recursive Functions of Symbolic Expressions and Their Computation by Machine, Part I
17. Foundations for structured programming with GADTs
18. Universal coalgebra: a theory of systems
19. Dependent Types in Practical Programming
20. The essence of compiling with continuations
21. Categories for the Working Mathematician (Graduate Texts in Mathematics)
22. A {S}yntactic {T}heory of {D}ynamic {B}inding
23. The ontology of biological sequences.
24. Additional gene ontology structure for improved biological reasoning.
25. Domain specific embedded compilers
26. Abstract interpretation: a unified lattice model for static analysis of programs by construction or approximation of fixpoints
27. Type Classes with Functional Dependencies
28. A Tutorial on (Co)Algebras and (Co)Induction
29. Languages of the Future
30. Macros as multi-stage computations: type-safe, generative, binding macros in MacroML
31. Categories for the Working Mathematician
32. Types, abstraction and parametric polymorphism
33. The categorical abstract machine
34. Monadic Parsing in Haskell
35. A relation based measure of semantic similarity for Gene Ontology annotations
36. Representing Control: A study of the CPS transformation
37. The design and implementation of typed scheme
38. Cloning-based context-sensitive pointer alias analysis using binary decision diagrams
39. Monadic Parser Combinators
40. Lectures on the Curry-Howard isomorphism
41. The {Calculus of Constructions}
42. Dynamic optimization for functional reactive programming using generalized algebraic data types
43. Fast and Loose Reasoning Is Morally Correct
44. Scrap your boilerplate: a practical design pattern for generic programming
45. Ownership types for safe programming: preventing data races and deadlocks
46. Parsing expression grammars: a recognition-based syntactic foundation
47. Finger trees: a simple general-purpose data structure
48. Adaptive Functional Programming
49. Definitional interpreters for higher-order programming languages
50. A new notation for arrows
For the CTPF models, you may access the raw topic distributions by computing,
model.alef ./ model.bet
Raw scores, as well as document and user recommendations, may be accessed via,
model.scores
### M x U matrix
### M = number of documents, ordered identically to the documents in model.corp.docs.
### U = number of users, ordered identically to the keys in model.corp.users.
model.drecs
model.urecs
Note, as was done by Blei et al. in their original paper, if you would like to warm start your CTPF model using the topic distributions generated by one of the other models, simply do the following prior to training your model,
ctpf_model.alef = exp.(model.beta)
### For model of type: LDA, fLDA, CTM, fCTM, gpuLDA, gpuCTM.
GPU Acceleration
GPU accelerating your model runs its performance bottlenecks on the GPU.
There's no reason to instantiate GPU models directly, instead you can simply instantiate the normal version of a supported model, and then use the @gpu macro to train it on the GPU,
model = LDA(readcorp(:nsf), 20)
@gpu train!(model, checkelbo=Inf)
### training...
Important. Notice that we did not check the ELBO at all during training. While you may check the ELBO if you wish, it's recommended that you do so infrequently, as computing the ELBO is done entirely on the CPU.
Here are the log scale benchmarks of the coordinate ascent algorithms for the GPU models, compared against their CPU equivalents,
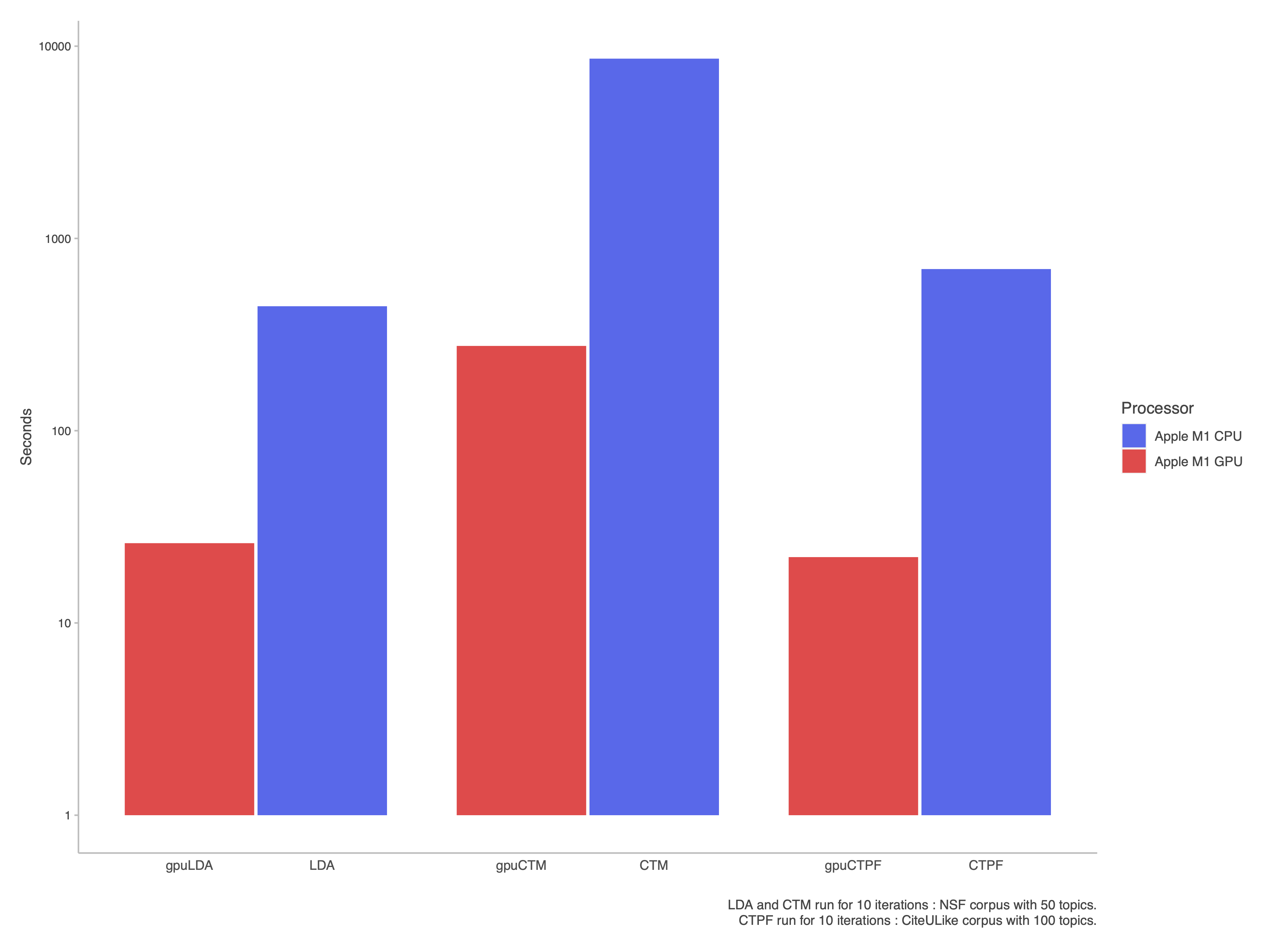
As we can see, running your model on the GPU is significantly faster than running it on the CPU.
Note that it's expected that your computer will lag when training on the GPU, since you're effectively siphoning off its rendering resources to fit your model.
Glossary
Types
mutable struct Document
"Document mutable struct"
"terms: A vector{Int} containing keys for the Corpus vocab Dict."
"counts: A Vector{Int} denoting the counts of each term in the Document."
"readers: A Vector{Int} denoting the keys for the Corpus users Dict."
"ratings: A Vector{Int} denoting the ratings for each reader in the Document."
"title: The title of the document (String)."
terms::Vector{Int}
counts::Vector{Int}
readers::Vector{Int}
ratings::Vector{Int}
title::String
mutable struct Corpus
"Corpus mutable struct."
"docs: A Vector{Document} containing the documents which belong to the Corpus."
"vocab: A Dict{Int, String} containing a mapping term Int (key) => term String (value)."
"users: A Dict{Int, String} containing a mapping user Int (key) => user String (value)."
docs::Vector{Document}
vocab::Dict{Int, String}
users::Dict{Int, String}
abstract type TopicModel end
mutable struct LDA <: TopicModel
"LDA mutable struct."
corpus::Corpus
K::Int
...
mutable struct fLDA <: TopicModel
"fLDA mutable struct."
corpus::Corpus
K::Int
...
mutable struct CTM <: TopicModel
"CTM mutable struct."
corpus::Corpus
K::Int
...
mutable struct fCTM <: TopicModel
"fCTM mutable struct."
corpus::Corpus
K::Int
...
mutable struct CTPF <: TopicModel
"CTPF mutable struct."
corpus::Corpus
K::Int
...
mutable struct gpuLDA <: TopicModel
"gpuLDA mutable struct."
corpus::Corpus
K::Int
...
mutable struct gpuCTM <: TopicModel
"gpuCTM mutable struct."
corpus::Corpus
K::Int
...
mutable struct gpuCTPF <: TopicModel
"gpuCTPF mutable struct."
corpus::Corpus
K::Int
...
Document/Corpus Functions
function check_doc(doc::Document)
"Check Document parameters."
function check_corp(corp::Corpus)
"Check Corpus parameters."
function readcorp(;docfile::String="", vocabfile::String="", userfile::String="", titlefile::String="", delim::Char=',', counts::Bool=false, readers::Bool=false, ratings::Bool=false)
"Load a Corpus object from text file(s)."
### readcorp(:nsf) - National Science Foundation Corpus.
### readcorp(:citeu) - CiteULike Corpus.
function writecorp(corp::Corpus; docfile::String="", vocabfile::String="", userfile::String="", titlefile::String="", delim::Char=',', counts::Bool=false, readers::Bool=false, ratings::Bool=false)
"Write a corpus."
function abridge_corp!(corp::Corpus, n::Integer=0)
"All terms which appear less than n times in the corpus are removed from all documents."
function alphabetize_corp!(corp::Corpus; vocab::Bool=true, users::Bool=true)
"Alphabetize vocab and/or user dictionaries."
function remove_terms!(corp::Corpus; terms::Vector{String}=[])
"Vocab keys for specified terms are removed from all documents."
function compact_corp!(corp::Corpus; vocab::Bool=true, users::Bool=true)
"Relabel vocab and/or user keys so that they form a unit range."
function condense_corp!(corp::Corpus)
"Ignore term order in documents."
"Multiple seperate occurrences of terms are stacked and their associated counts increased."
function pad_corp!(corp::Corpus; vocab::Bool=true, users::Bool=true)
"Enter generic values for vocab and/or user keys which appear in documents but not in the vocab/user dictionaries."
function remove_empty_docs!(corp::Corpus)
"Documents with no terms are removed from the corpus."
function remove_redundant!(corp::Corpus; vocab::Bool=true, users::Bool=true)
"Remove vocab and/or user keys which map to redundant values."
"Reassign Document term and/or reader keys."
function stop_corp!(corp::Corpus)
"Filter stop words in the associated corpus."
function trim_corp!(corp::Corpus; vocab::Bool=true, users::Bool=true)
"Those keys which appear in the corpus vocab and/or user dictionaries but not in any of the documents are removed from the corpus."
function trim_docs!(corp::Corpus; terms::Bool=true, readers::Bool=true)
"Those vocab and/or user keys which appear in documents but not in the corpus dictionaries are removed from the documents."
function fixcorp!(corp::Corpus; vocab::Bool=true, users::Bool=true, abridge::Integer=0, alphabetize::Bool=false, condense::Bool=false, pad::Bool=false, remove_empty_docs::Bool=false, remove_redundant::Bool=false, remove_terms::Vector{String}=String[], stop::Bool=false, trim::Bool=false)
"Generic function to ensure that a Corpus object can be loaded into a TopicModel object."
"Either pad_corp! or trim_docs!."
"compact_corp!."
"Contains other optional keyword arguments."
function showdocs(corp::Corpus, docs / doc_indices)
"Display document(s) in readable format."
function showtitles(corp::Corpus, docs / doc_indices)
"Display document title(s) in readable format."
function getvocab(corp::Corpus)
function getusers(corp::Corpus)
Model Functions
function showdocs(model::TopicModel, docs / doc_indices)
"Display document(s) in readable format."
function showtitles(model::TopicModel, docs / doc_indices)
"Display document title(s) in readable format."
function check_model(model::TopicModel)
"Check model parameters."
function train!(model::TopicModel; iter::Integer=150, tol::Real=1.0, niter::Integer=1000, ntol::Real=1/model.K^2, viter::Integer=10, vtol::Real=1/model.K^2, checkelbo::Union{Integer, Inf}=1, printelbo::Bool=true)
"Train TopicModel."
### 'iter' - maximum number of iterations through the corpus.
### 'tol' - absolute tolerance for ∆elbo as a stopping criterion.
### 'niter' - maximum number of iterations for Newton's and interior-point Newton's methods. (not included for CTPF and gpuCTPF models.)
### 'ntol' - tolerance for change in function value as a stopping criterion for Newton's and interior-point Newton's methods. (not included for CTPF and gpuCTPF models.)
### 'viter' - maximum number of iterations for optimizing variational parameters (at the document level).
### 'vtol' - tolerance for change in variational parameter values as stopping criterion.
### 'checkelbo' - number of iterations between ∆elbo checks (for both evaluation and convergence of the evidence lower-bound).
### 'printelbo' - if true, print ∆elbo to REPL.
@gpu train!
"Train model on GPU."
function gendoc(model::TopicModel, laplace_smooth::Real=0.0)
"Generate a generic document from model parameters by running the associated graphical model as a generative process."
function gencorp(model::TopicModel, M::Integer, laplace_smooth::Real=0.0)
"Generate a generic corpus of size M from model parameters."
function showtopics(model::TopicModel, V::Integer=15; topics::Union{Integer, Vector{<:Integer}, UnitRange{<:Integer}}=1:model.K, cols::Integer=4)
"Display the top V words for each topic in topics."
function showlibs(model::Union{CTPF, gpuCTPF}, users::Union{Integer, Vector{<:Integer}, UnitRange{<:Integer}})
"Show the document(s) in a user's library."
function showdrecs(model::Union{CTPF, gpuCTPF}, docs::Union{Integer, Vector{<:Integer}, UnitRange{<:Integer}}, U::Integer=16; cols=4)
"Show the top U user recommendations for a document(s)."
function showurecs(model::Union{CTPF, gpuCTPF}, users::Union{Integer, Vector{<:Integer}, UnitRange{<:Integer}}, M::Integer=10; cols=1)
"Show the top M document recommendations for a user(s)."
function predict(corp::Corpus, train_model::Union{LDA, gpuLDA, fLDA, CTM, gpuCTM, fCTM}; iter::Integer=10, tol::Real=1/train_model.K^2, niter::Integer=1000, ntol::Real=1/train_model.K^2)
"Predict topic distributions for corpus of documents based on trained LDA or CTM model."
function topicdist(model::TopicModel, doc_indices::Union{Integer, Vector{<:Integer}, UnitRange{<:Integer}})
"Get TopicModel topic distributions for document(s) as a probability vector."
Bibliography
- Latent Dirichlet Allocation (2003); Blei, Ng, Jordan. pdf
- Filtered Latent Dirichlet Allocation: Variational Bayes Algorithm (2016); Proffitt. pdf
- Correlated Topic Models (2006); Blei, Lafferty. pdf
- Content-based Recommendations with Poisson Factorization (2014); Gopalan, Charlin, Blei. pdf
- Numerical Optimization (2006); Nocedal, Wright. Amazon
- Machine Learning: A Probabilistic Perspective (2012); Murphy. Amazon
- OpenCL in Action: How to Accelerate Graphics and Computation (2011); Scarpino. Amazon