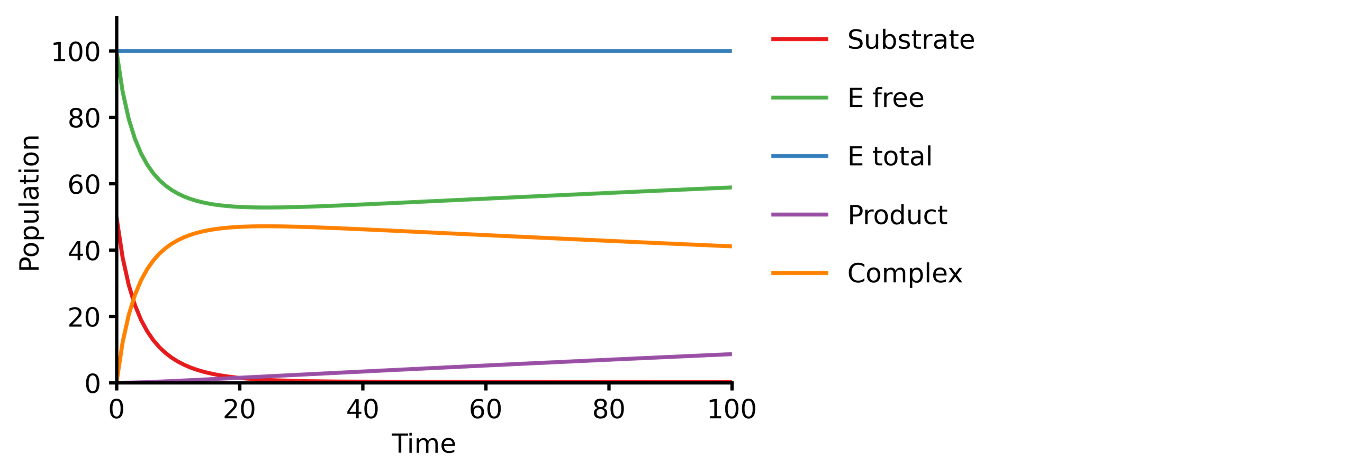Model Construction
pasmopy.Text2Model allows you to build a BioMASS model from text. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model.
Example
This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model with Pasmopy.
E + S ⇄ ES → E + P
An enzyme, E, binding to a substrate, S, to form a complex, ES, which in turn releases a product, P, regenerating the original enzyme.
Prepare a text file describing biochemical reactions (e.g.,
michaelis_menten.txt)``` E binds S <–> ES | kf=0.003, kr=0.001 | E=100, S=50 ES dissociates to E and P | kf=0.002, kr=0
@obs Substrate: u[S] @obs Efree: u[E] @obs Etotal: u[E] + u[ES] @obs Product: u[P] @obs Complex: u[ES]
@sim tspan: [0, 100] ```
Convert the text into an executable model
shell $ python```pythonfrom pasmopy import Text2Model description = Text2Model("michaelismenten.txt", lang="julia") description.convert() # generate 'michaelismenten_jl/'
Model information
2 reactions 4 species 4 parameters ```
Run simulation
shell $ julia```julia using BioMASSmodel = Model("./michaelismentenjl"); run_simulation(model) ```