Bacterial Growth
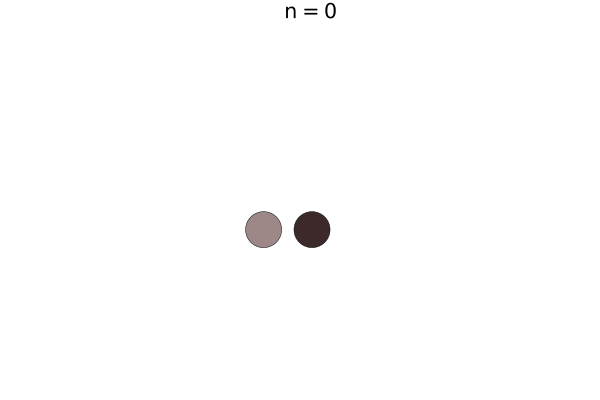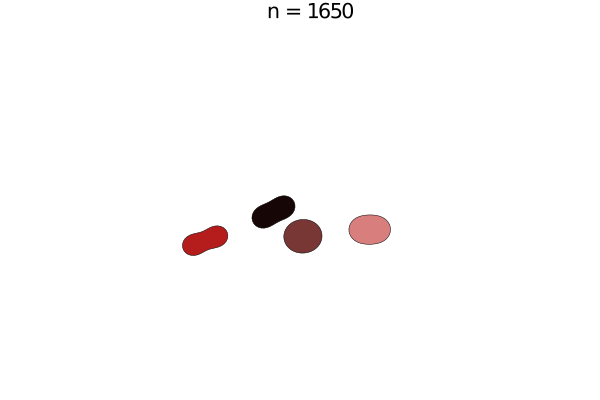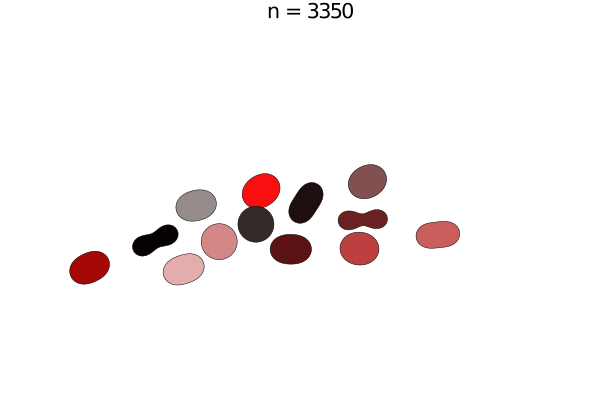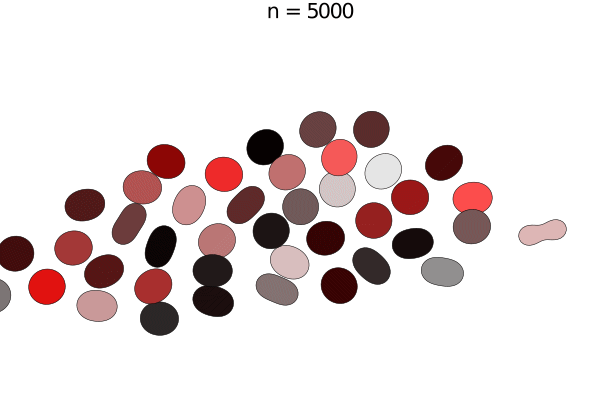
Bacterial colonies are a prime example for growing active matter, where systems are driven out of equilibrium by proliferation. This model is a simplified version of unpublished work by Yoav G. Pollack and Philip Bittihn; similar models can be found in literature. Here, a bacterium is modelled by two soft disk "nodes" linked by a spring, whose rest length grows with a constant growth rate. When it has reached its full extension, the cell divides into two daughter cells with the same orientation.
This example is a showcase of a complex continuous system. Agents will be splitting into more agents, thus having agent generation in continuous space. The model also uses advanced agent movement in continuous space, where a specialized "move_agent" function is created. Advanced plotting is also done, since each agent is a specialized shape. It is also available from the Models module as Models.growing_bacteria.
using Agents, LinearAlgebra
mutable struct SimpleCell <: AbstractAgent
id::Int
pos::NTuple{2,Float64}
length::Float64
orientation::Float64
growthprog::Float64
growthrate::Float64
# node positions/forces
p1::NTuple{2,Float64}
p2::NTuple{2,Float64}
f1::NTuple{2,Float64}
f2::NTuple{2,Float64}
end
function SimpleCell(id, pos, l, φ, g, γ)
a = SimpleCell(id, pos, l, φ, g, γ, (0.0, 0.0), (0.0, 0.0), (0.0, 0.0), (0.0, 0.0))
update_nodes!(a)
return a
endMain.ex-growing_bacteria.SimpleCell
In this model, the agents have to store their state in two redundant ways: the cell coordinates (position, length, orientation) are required for the equations of motion, while the positions of the disk-shaped nodes are necessary for calculating mechanical forces between cells. To transform from one set of coordinates to the other, we need to write a function
function update_nodes!(a::SimpleCell)
offset = 0.5 * a.length .* unitvector(a.orientation)
a.p1 = a.pos .+ offset
a.p2 = a.pos .- offset
endSome geometry convenience functions
unitvector(φ) = reverse(sincos(φ))
cross2D(a, b) = a[1] * b[2] - a[2] * b[1]Stepping functions
function model_step!(model)
for a in allagents(model)
if a.growthprog ≥ 1
# When a cell has matured, it divides into two daughter cells on the
# positions of its nodes.
add_agent!(a.p1, model, 0.0, a.orientation, 0.0, 0.1 * rand() + 0.05)
add_agent!(a.p2, model, 0.0, a.orientation, 0.0, 0.1 * rand() + 0.05)
kill_agent!(a, model)
else
# The rest lengh of the internal spring grows with time. This causes
# the nodes to physically separate.
uv = unitvector(a.orientation)
internalforce = model.hardness * (a.length - a.growthprog) .* uv
a.f1 = -1 .* internalforce
a.f2 = internalforce
end
end
# Bacteria can interact with more than on other cell at the same time, therefore,
# we need to specify the option `:all` in `interacting_pairs`
for (a1, a2) in interacting_pairs(model, 2.0, :all)
interact!(a1, a2, model)
end
endHere we use a custom move_agent! function, because the agents have several moving parts. Notice that the first derivatives of all degrees of freedom is directly proportional to the force applied to them. This overdamped approximation is valid for small length scales, where viscous forces dominate over inertia.
function agent_step!(agent::SimpleCell, model::ABM)
fsym, compression, torque = transform_forces(agent)
agent.pos = agent.pos .+ model.dt * model.mobility .* fsym
agent.length += model.dt * model.mobility .* compression
agent.orientation += model.dt * model.mobility .* torque
agent.growthprog += model.dt * agent.growthrate
update_nodes!(agent)
update_space!(model, agent)
return agent.pos
endHelper functions
function interact!(a1::SimpleCell, a2::SimpleCell, model)
n11 = noderepulsion(a1.p1, a2.p1, model)
n12 = noderepulsion(a1.p1, a2.p2, model)
n21 = noderepulsion(a1.p2, a2.p1, model)
n22 = noderepulsion(a1.p2, a2.p2, model)
a1.f1 = @. a1.f1 + (n11 + n12)
a1.f2 = @. a1.f2 + (n21 + n22)
a2.f1 = @. a2.f1 - (n11 + n21)
a2.f2 = @. a2.f2 - (n12 + n22)
end
function noderepulsion(p1::NTuple{2,Float64}, p2::NTuple{2,Float64}, model::ABM)
delta = p1 .- p2
distance = norm(delta)
if distance ≤ 1
uv = delta ./ distance
return (model.hardness * (1 - distance)) .* uv
end
return (0, 0)
end
function transform_forces(agent::SimpleCell)
# symmetric forces (CM movement)
fsym = agent.f1 .+ agent.f2
# antisymmetric forces (compression, torque)
fasym = agent.f1 .- agent.f2
uv = unitvector(agent.orientation)
compression = dot(uv, fasym)
torque = 0.5 * cross2D(uv, fasym)
return fsym, compression, torque
endAnimating bacterial growth
Okay, we can now initialize a model and see what it does.
space = ContinuousSpace(2, extend = (12, 12), periodic = false, metric = :euclidean)
model = ABM(
SimpleCell,
space,
properties = Dict(:dt => 0.005, :hardness => 1e2, :mobility => 1.0),
)AgentBasedModel with 0 agents of type SimpleCell space: 2-dimensional ContinuousSpace scheduler: fastest properties: Dict(:hardness => 100.0,:dt => 0.005,:mobility => 1.0)
Let's start with just two agents.
add_agent!((5.0, 5.0), model, 0.0, 0.3, 0.0, 0.1)
add_agent!((6.0, 5.0), model, 0.0, 0.0, 0.0, 0.1)The model has several parameters, and some of them are of interest. We could e.g. define
adata = [:pos, :length, :orientation, :growthprog, :p1, :p2, :f1, :f2]and then run! the model. But we'll animate the model directly.
Here we once again use the huge flexibility provided by plotabm to plot the becteria cells. We define a function that creates a custom Shape based on the agent:
using AgentsPlots
function cassini_oval(agent)
t = LinRange(0, 2π, 50)
a = agent.growthprog
b = 1
m = @. 2 * sqrt((b^4 - a^4) + a^4 * cos(2 * t)^2) + 2 * a^2 * cos(2 * t)
C = sqrt.(m / 2)
x = C .* cos.(t)
y = C .* sin.(t)
uv = reverse(sincos(agent.orientation))
θ = atan(uv[2], uv[1])
R = [cos(θ) -sin(θ); sin(θ) cos(θ)]
bacteria = R * permutedims([x y])
Shape(bacteria[1, :], bacteria[2, :])
endset up some nice colors
bacteria_colors(agent) =
HSV.(agent.id * 2.718 .% 1, agent.id * 3.14 .% 1, agent.id * 1.618 .% 1)and proceed with the animation
e = model.space.extend
anim = @animate for i in 0:50:5000
step!(model, agent_step!, model_step!, 100)
p1 = plotabm(
model,
am = cassini_oval,
as = 30,
ac = bacteria_colors,
showaxis = false,
grid = false,
xlims = (0, e[1]),
ylims = (0, e[2]),
)
title!(p1, "n = $(i)")
end
gif(anim, "bacteria.gif", fps = 25)