SIR model for the spread of COVID-19
 This example illustrates how to use
This example illustrates how to use GraphSpace and how to model agents on an graph (network) where the transition probabilities between each node is not constant.
SIR model
A SIR model tracks the ratio of Susceptible, Infected, and Recovered individuals within a population. Here we add one more category of individuals: those who are infected, but do not know it. Transmission rate for infected and diagnosed individuals is lower than infected and undetected. We also allow a fraction of recovered individuals to catch the disease again, meaning that recovering the disease does not bring full immunity.
Model parameters
Here are the model parameters, some of which have default values.
Ns: a vector of population sizes per city. The amount of cities is justC=length(Ns).β_und: a vector for transmission probabilities β of the infected but undetected per city. Transmission probability is how many susceptible are infected per day by an infected individual. If social distancing is practiced, this number increases.β_det: an array for transmission probabilities β of the infected and detected per city. If hospitals are full, this number increases.infection_period = 30: how many days before a person dies or recovers.detection_time = 14: how many days before an infected person is detected.death_rate = 0.02: the probability that the individual will die after theinfection_period.reinfection_probability = 0.05: The probability that a recovered person can get infected again.migration_rates: A matrix of migration probability per individual per day from one city to another.Is = [zeros(C-1)..., 1]: An array for initial number of infected but undetected people per city. This starts as only one infected individual in the last city.
Notice that Ns, β, Is all need to have the same length, as they are numbers for each city. We've tried to add values to the infection parameters similar to the ones you would hear on the news about COVID-19.
The good thing with Agent based models is that you could easily extend the model we implement here to also include age as an additional property of each agent. This makes ABMs flexible and suitable for research of virus spreading.
Making the model in Agents.jl
We start by defining the PoorSoul agent type and the ABM
using Agents, Random, DataFrames, LightGraphs
using Distributions: Poisson, DiscreteNonParametric
using DrWatson: @dict
using Plots
mutable struct PoorSoul <: AbstractAgent
id::Int
pos::Int
days_infected::Int # number of days since is infected
status::Symbol # 1: S, 2: I, 3:R
end
function model_initiation(;
Ns,
migration_rates,
β_und,
β_det,
infection_period = 30,
reinfection_probability = 0.05,
detection_time = 14,
death_rate = 0.02,
Is = [zeros(Int, length(Ns) - 1)..., 1],
seed = 0,
)
Random.seed!(seed)
@assert length(Ns) ==
length(Is) ==
length(β_und) ==
length(β_det) ==
size(migration_rates, 1) "length of Ns, Is, and B, and number of rows/columns in migration_rates should be the same "
@assert size(migration_rates, 1) == size(migration_rates, 2) "migration_rates rates should be a square matrix"
C = length(Ns)
# normalize migration_rates
migration_rates_sum = sum(migration_rates, dims = 2)
for c in 1:C
migration_rates[c, :] ./= migration_rates_sum[c]
end
properties = @dict(
Ns,
Is,
β_und,
β_det,
β_det,
migration_rates,
infection_period,
infection_period,
reinfection_probability,
detection_time,
C,
death_rate
)
space = GraphSpace(complete_digraph(C))
model = ABM(PoorSoul, space; properties = properties)
# Add initial individuals
for city in 1:C, n in 1:Ns[city]
ind = add_agent!(city, model, 0, :S) # Susceptible
end
# add infected individuals
for city in 1:C
inds = get_node_contents(city, model)
for n in 1:Is[city]
agent = model[inds[n]]
agent.status = :I # Infected
agent.days_infected = 1
end
end
return model
endWe will make a function that starts a model with C number of cities, and creates the other parameters automatically by attributing some random values to them. You could directly use the above constructor and specify all Ns, β, etc. for a given set of cities.
All cities are connected with each other, while it is more probable to travel from a city with small population into a city with large population.
using LinearAlgebra: diagind
function create_params(;
C,
max_travel_rate,
infection_period = 30,
reinfection_probability = 0.05,
detection_time = 14,
death_rate = 0.02,
Is = [zeros(Int, C - 1)..., 1],
seed = 19,
)
Random.seed!(seed)
Ns = rand(50:5000, C)
β_und = rand(0.3:0.02:0.6, C)
β_det = β_und ./ 10
Random.seed!(seed)
migration_rates = zeros(C, C)
for c in 1:C
for c2 in 1:C
migration_rates[c, c2] = (Ns[c] + Ns[c2]) / Ns[c]
end
end
maxM = maximum(migration_rates)
migration_rates = (migration_rates .* max_travel_rate) ./ maxM
migration_rates[diagind(migration_rates)] .= 1.0
params = @dict(
Ns,
β_und,
β_det,
migration_rates,
infection_period,
reinfection_probability,
detection_time,
death_rate,
Is
)
return params
end
params = create_params(C = 8, max_travel_rate = 0.01)
model = model_initiation(; params...)AgentBasedModel with 17319 agents of type PoorSoul
space: GraphSpace with 8 nodes and 56 edges
scheduler: fastest
properties: Dict{Symbol,Any}(:Is => [0, 0, 0, 0, 0, 0, 0, 1],:death_rate => 0.02,:infection_period => 30,:β_und => [0.44, 0.58, 0.34, 0.46, 0.32, 0.6, 0.48, 0.6],:Ns => [3154, 630, 2351, 1597, 3910, 635, 193, 4849],:migration_rates => [0.9956206383765343 0.00045723324181758666 … 0.0004044290857197312 0.0009670289731147321; 0.002270036979920437 0.9873426241439373 … 0.0004937210450514058 0.003286874369181837; … ; 0.006403572378352215 0.0015745862167265829 … 0.9646492958366257 0.00964649295836626; 0.0006294580658856994 0.0004309384909393661 … 0.00039656723331197 0.9963494892900222],:detection_time => 14,:reinfection_probability => 0.05,:β_det => [0.044, 0.057999999999999996, 0.034, 0.046, 0.032, 0.06, 0.048, 0.06],:C => 8…)Alright, let's plot the cities as a graph to get an idea how the model "looks like", using the function plotabm.
using AgentsPlots
plotargs = (node_size = 0.2, method = :circular, linealpha = 0.4)
plotabm(model; plotargs...)The node size is proportional to the relative population of each city. In principle we could adjust the edge widths to be proportional with the migration rates, by doing:
g = model.space.graph
edgewidthsdict = Dict()
for node in 1:nv(g)
nbs = neighbors(g, node)
for nb in nbs
edgewidthsdict[(node, nb)] = params[:migration_rates][node, nb]
end
end
edgewidthsf(s, d, w) = edgewidthsdict[(s, d)] * 250
plotargs = merge(plotargs, (edgewidth = edgewidthsf,))
plotabm(model; plotargs...)In the following we will be coloring each node according to how large percentage of the population is infected. So we create a function to give to plotabm as a second argument
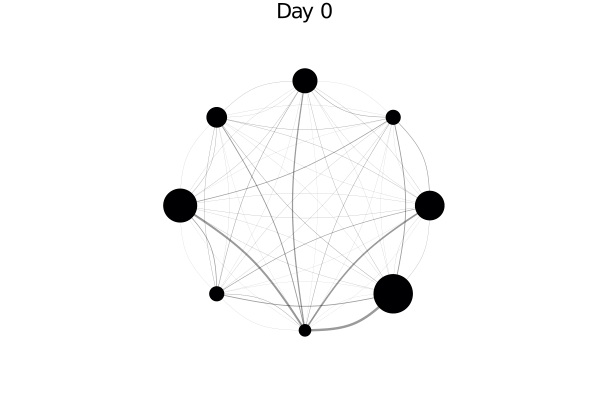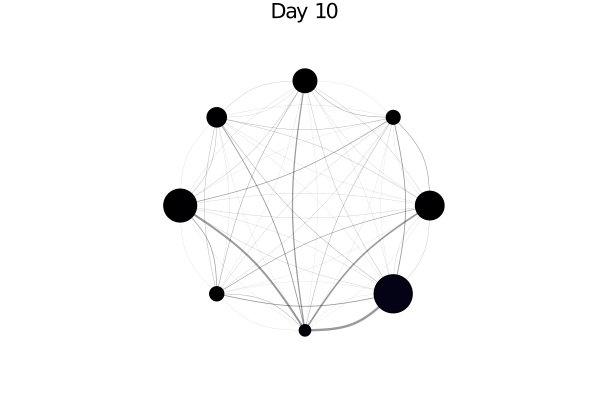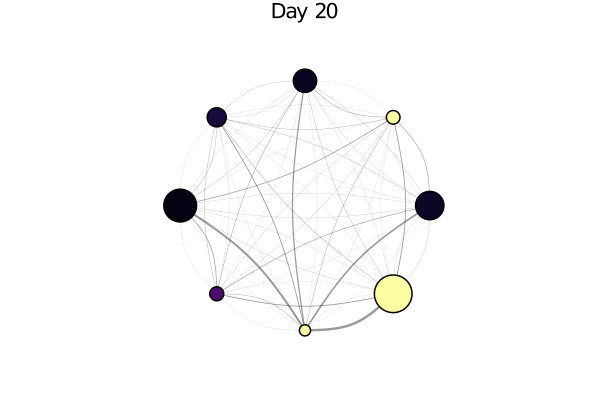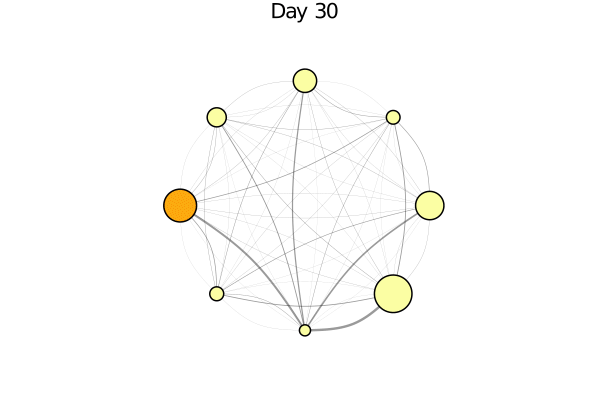
infected_fraction(x) = cgrad(:inferno)[count(a.status == :I for a in x) / length(x)]
plotabm(model; ac = infected_fraction, plotargs...)Here this shows all nodes as black, since we haven't run the model yet. Let's change that!
SIR Stepping functions
Now we define the functions for modelling the virus spread in time
function agent_step!(agent, model)
migrate!(agent, model)
transmit!(agent, model)
update!(agent, model)
recover_or_die!(agent, model)
end
function migrate!(agent, model)
nodeid = agent.pos
d = DiscreteNonParametric(1:(model.C), model.migration_rates[nodeid, :])
m = rand(d)
if m ≠ nodeid
move_agent!(agent, m, model)
end
end
function transmit!(agent, model)
agent.status == :S && return
rate = if agent.days_infected < model.detection_time
model.β_und[agent.pos]
else
model.β_det[agent.pos]
end
d = Poisson(rate)
n = rand(d)
n == 0 && return
for contactID in get_node_contents(agent, model)
contact = model[contactID]
if contact.status == :S ||
(contact.status == :R && rand() ≤ model.reinfection_probability)
contact.status = :I
n -= 1
n == 0 && return
end
end
end
update!(agent, model) = agent.status == :I && (agent.days_infected += 1)
function recover_or_die!(agent, model)
if agent.days_infected ≥ model.infection_period
if rand() ≤ model.death_rate
kill_agent!(agent, model)
else
agent.status = :R
agent.days_infected = 0
end
end
endExample animation
model = model_initiation(; params...)
anim = @animate for i in 0:30
i > 0 && step!(model, agent_step!, 1)
p1 = plotabm(model; ac = infected_fraction, plotargs...)
title!(p1, "Day $(i)")
end
gif(anim, "covid_evolution.gif", fps = 5)One can really see "explosive growth" in this animation. Things look quite calm for a while and then suddenly supermarkets have no toilet paper anymore!
Exponential growth
We now run the model and collect data. We define two useful functions for data collection:
infected(x) = count(i == :I for i in x)
recovered(x) = count(i == :R for i in x)and then collect data
model = model_initiation(; params...)
to_collect = [(:status, f) for f in (infected, recovered, length)]
data, _ = run!(model, agent_step!, 100; adata = to_collect)
data[1:10, :]10 rows × 4 columns
| step | infected_status | recovered_status | length_status | |
|---|---|---|---|---|
| Int64 | Int64 | Int64 | Int64 | |
| 1 | 0 | 1 | 0 | 17319 |
| 2 | 1 | 2 | 0 | 17319 |
| 3 | 2 | 3 | 0 | 17319 |
| 4 | 3 | 11 | 0 | 17319 |
| 5 | 4 | 18 | 0 | 17319 |
| 6 | 5 | 31 | 0 | 17319 |
| 7 | 6 | 69 | 0 | 17319 |
| 8 | 7 | 116 | 0 | 17319 |
| 9 | 8 | 217 | 0 | 17319 |
| 10 | 9 | 383 | 0 | 17319 |
We now plot how quantities evolved in time to show the exponential growth of the virus
N = sum(model.Ns) # Total initial population
x = data.step
p = plot(
x,
log10.(data[:, aggname(:status, infected)]),
label = "infected",
xlabel = "steps",
ylabel = "log(count)",
)
plot!(p, x, log10.(data[:, aggname(:status, recovered)]), label = "recovered")
dead = log10.(N .- data[:, aggname(:status, length)])
plot!(p, x, dead, label = "dead")The exponential growth is clearly visible since the logarithm of the number of infected increases linearly, until everyone is infected.